Revolve cell fate choice with lineage tracing and single-cell genomics
A central interest of the Li lab is to understand how genetic and epigenetic regulators as well as cell niche interplay with each other, and how they collectively regulate cell identity and cell dynamics.
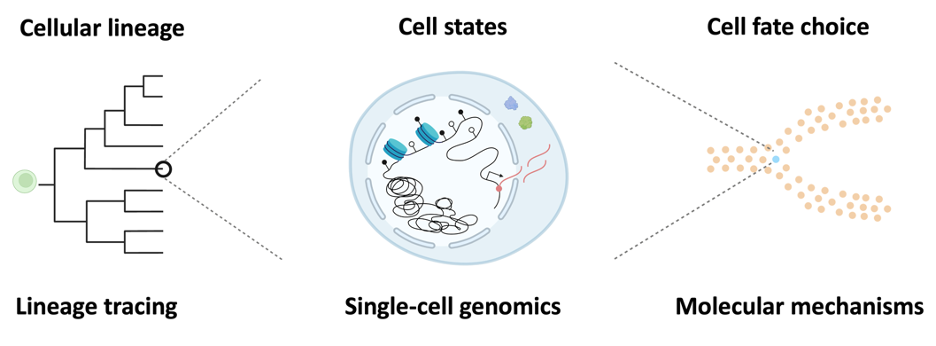
Recording cell fate choice
Let’s do genetic engineering!
High-resolution recording of cell fate choice will lay the sound foundation for the comprehensive understanding of this process. Dr. Li Li has developed DARLIN mouse model to systematically study the lineage relationships in various tissues under physiological or pathological conditions. The Li lab is currently using DARLIN to record cell fate commitment during embryo development, tissue regeneration and immune response. We are also developing next-generation lineage tracing tools to better visualize this process.
Decoding cell fate choice
Let’s do single-cell profiling!
We are interested in the epigenomic regulations during cell fate commitment. Dr. Li Li has developed Camellia-seq technology to systematically study the lineage relationships, transcriptome, and epigenome in single cells. Currently, we are applying Camellia-seq to study the epigenetic memory during embryo development. We are also developing epigenetic editors to better decode the underlying regulations in cell fate choice.
